

- #Cytoscape networks how to
- #Cytoscape networks update
- #Cytoscape networks software
- #Cytoscape networks download
- #Cytoscape networks free
Were responsible for the compound graph support.
#Cytoscape networks free
We appreciate people linking back to us, because that helps us keep Cytoscape Web development funded.įeel free to link to us however you choose or use one of the examples below:Ĭytoscape Web is maintained by Christian Lopes and Max Franz,įrom the Donnelly Centre for Cellular and Biomolecular Research.įrom the i-Vis Information Visualization Research Group It uses Cytoscape Web to display networks that illustrate the interactions among different resources. Pathguide is a meta-database that provides an overview of more thanģ00 web-accessible biological pathway and network databases. Of the interaction Reference Index (iRefIndex) which integrates protein interaction data from ten different interaction databases:īioGRID, BIND, CORUM, DIP, HPRD, INTACT, MINT, MPPI, MPACT and OPHID. IRefWeb is an interface to a relational database containing the latest build A lot of Apps are available for various kinds of problem domains, including bioinformatics, social network analysis, and semantic web.
#Cytoscape networks software
The rendering speed of Cytoscape Web allows for dynamic user Cytoscape is an open source software platform for visualizing complex networks and integrating these with any type of attribute data. The data provided to Cytoscape Web in GeneMANIA is unique to every user query, and easily Client applicationsĬytoscape Web is currently being used by a gene function prediction server, A Cytoscape App for multiple local alignment of protein-protein interaction (PPI) networks InsituNet Analysis of in situ gene expression data in terms of spatial co-expression. Once you upload your data, it should appear in two columns in the bottom right-hand corner of your screen. If you use Cytoscape Web, please link us back. We chose Flex/Flash because were not able to find any other solutions to the problem of visualizing interactive networks on the web that fit our requirements.Ĭytoscape Web is open-source software released under LGPL license. node shapes, visual mappings, XGMML support). Not as a complete user application, but as an independent component that could be reused by other developers.ĭespite using different technologies ( Flex and JavaScript)Ĭompared to Cytoscape, Cytoscape Web follows most of the Cytoscape concepts (e.g. The idea was to create a simple version of Cytoscape for the Web,
#Cytoscape networks how to
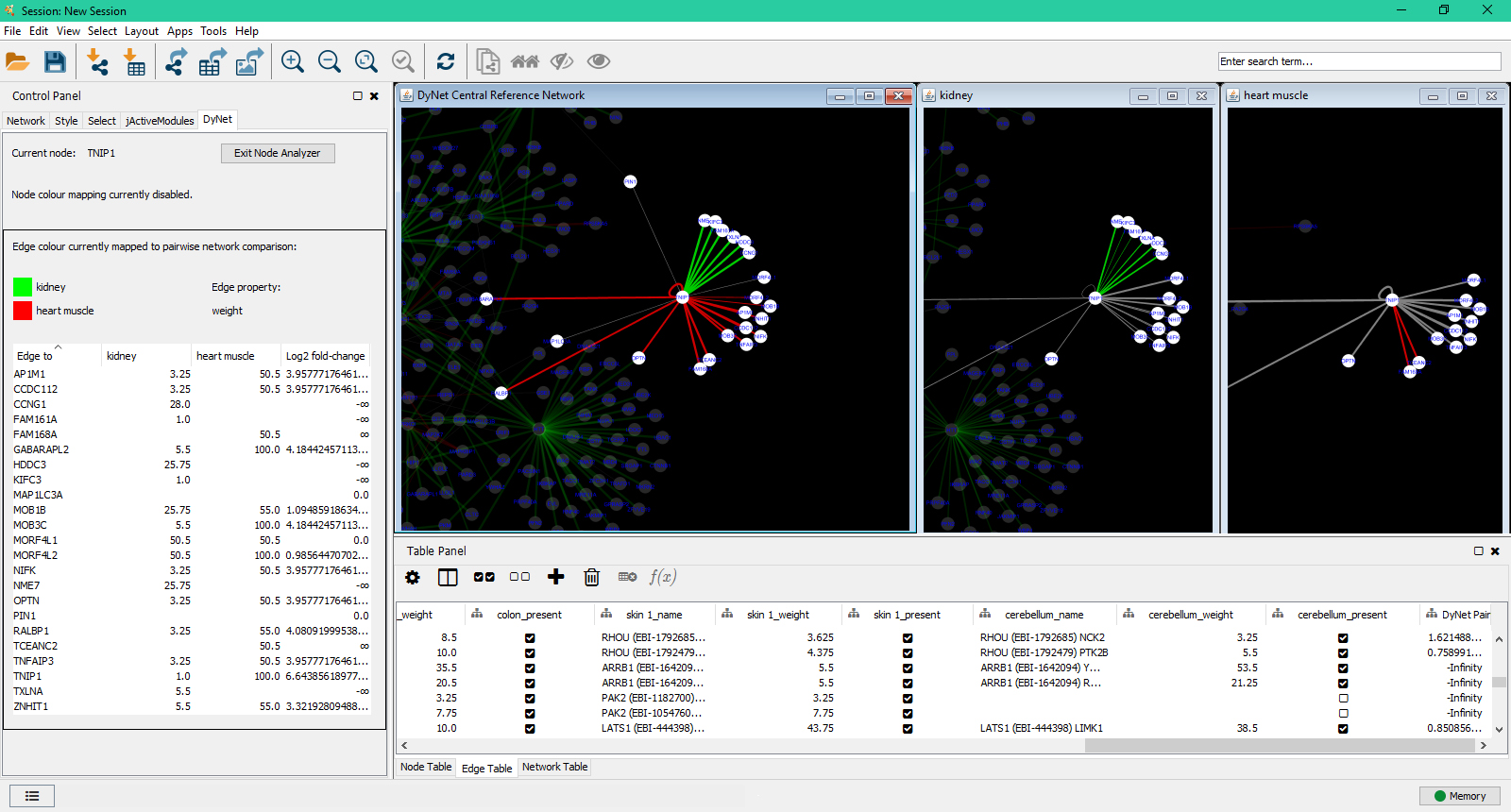
This tutorial outlines how to load network data from several popular sources and formats. Where you get your network data depends on your biological question and analysis plan. GeneMANIA project at the University of Toronto. Making Cytoscape Networks Homepage Making Cytoscape Networks ¶ For this visualization, we will use the real edge table ( realedgetable.txt) and real node file ( realnodetable.txt) that is generated by the makeotunetwork.py script. In Cytoscape, network data can be loaded from a variety of sources, and in several different formats. Including the Compound Spring Embedder layout algorithm.Ĭytoscape Web was created as a result of the need for a visualization interface for the Of Bilkent University implemented the compound graph feature,
The i-Vis Information Visualization Research Group National Institute of General Medical Sciences (NIGMS))Īnd number RR031228 (NIH National Center for Research Resources (NCRR) Biomedical Technology Research Center (BTRC)). It is now actively developed as part of the Cytoscape project which is funded by Ontario Genomics Institute (2007-OGI-TD-05). The Donnelly Centre for Cellular and Biomolecular Research,Ĭytoscape Web development was originally funded by Genome Canada, through the doi: 10.1186/gb-r80.Cytoscape Web was initially developed at the
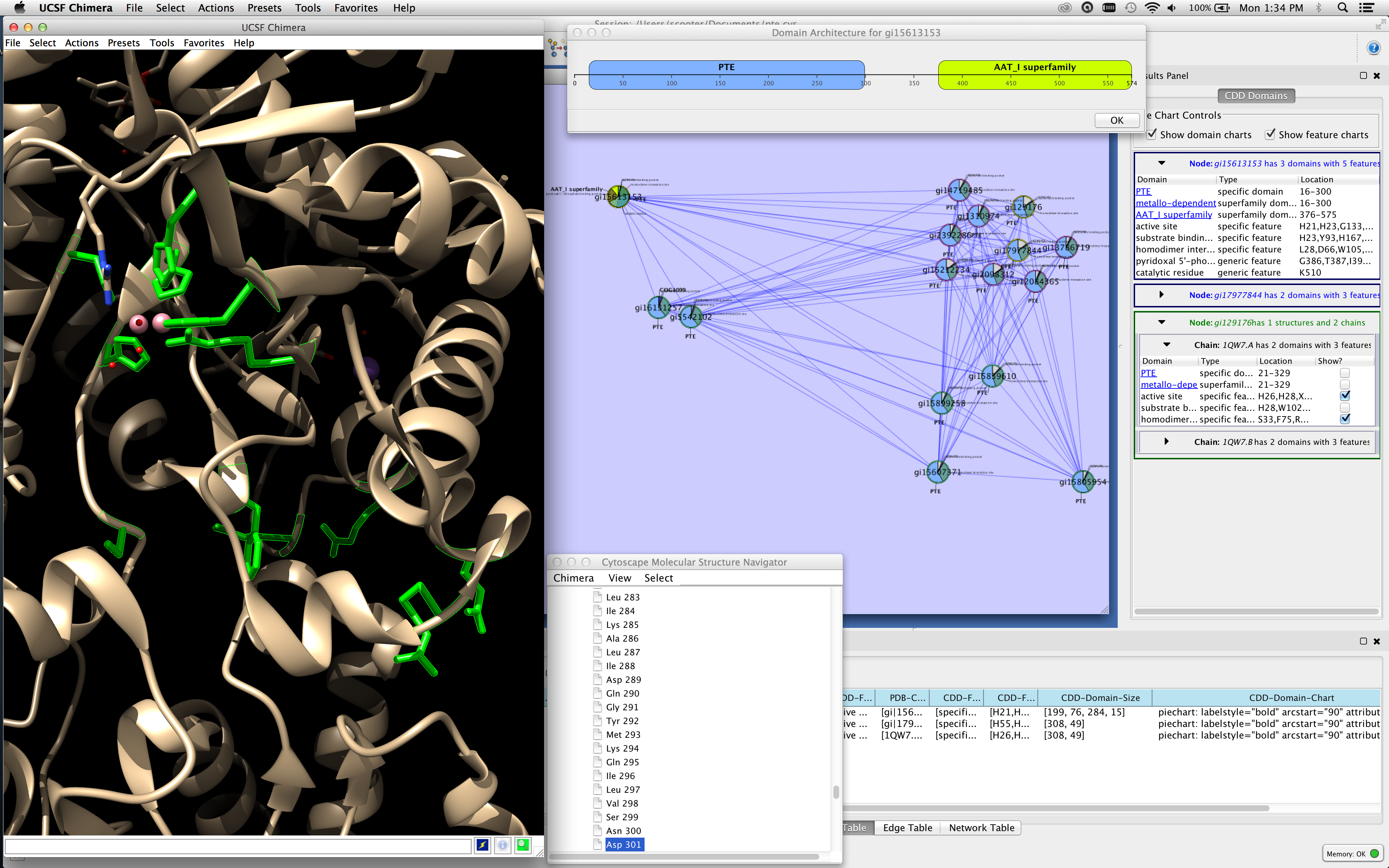
Bioconductor: open software development for computational biology and bioinformatics. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. The Taverna workflow suite: designing and executing workflows of Web Services on the desktop, web or in the cloud. Wolstencroft K, Haines R, Fellows D, Williams A, Withers D, Owen S, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. It is a bit verbose, and you may be able to omit some of the attributes depending on your needs: import networkx as nx G nx.
#Cytoscape networks update
Update Here is some code to add the graphics look-and-feel and positioning to a networkx graph.
#Cytoscape networks download
doi: 10.1101/gr.1239303.Īfgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, et al. If not, download it and drop it into your plugins folder, then restart Cytoscape and try loading the g.xml network again. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al.


 0 kommentar(er)
0 kommentar(er)
